Publications
Group highlights
At the end of this page, you can find the full list of publications. All papers are also available on Google Scholar.

We find that Estrogen-related receptors (ERR) signaling is necessary for induction of genes involved in mitochondrial and cardiac-specific contractile processes during human induced pluripotent stem cell-derived cardiomyocyte (hiPSC-CM) differentiation.
T. Sakamoto, K. Batmanov, S. Wan, Y. Guo, L. Lai, R. B. Vega and D. P. Kelly*.
Nature Communications 13, 1991 (2022)
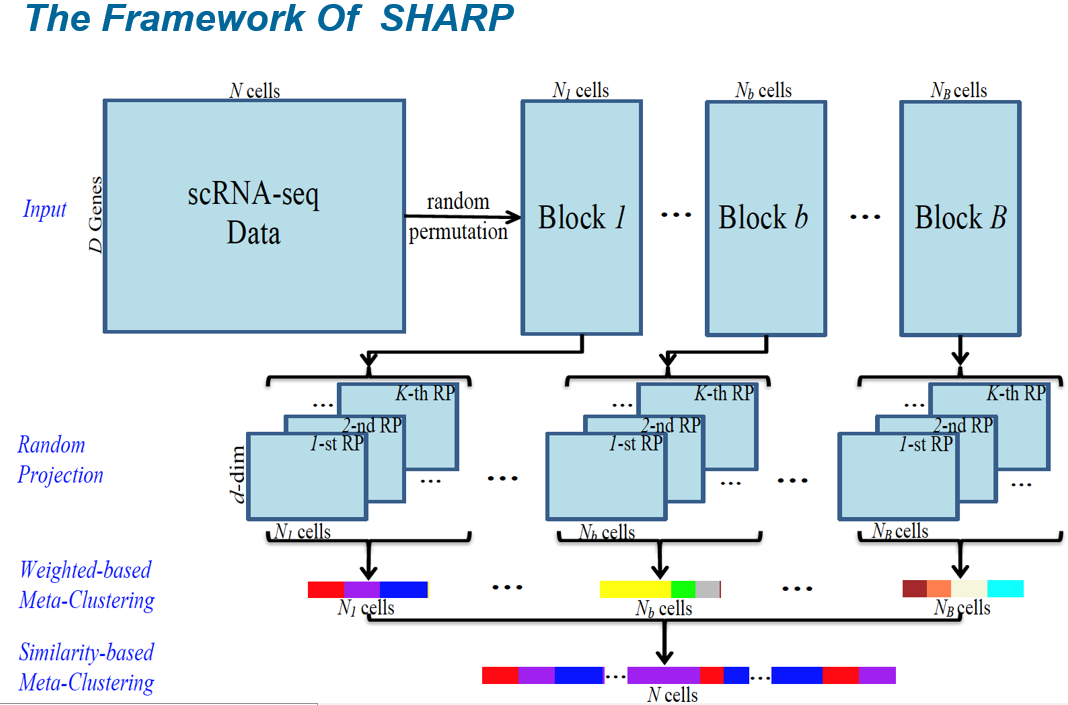
To process large-scale single-cell RNA-sequencing (scRNA-seq) data effectively without excessive distortion during dimension reduction, we present SHARP, an ensemble random projection-based algorithm that is scalable to clustering 10 million cells. Comprehensive benchmarking tests on 17 public scRNA-seq datasets demonstrate that SHARP outperforms existing methods in terms of speed and accuracy.
S. Wan, J. Kim and K. J. Won*.
Genome Research 30 (2), 205-213 (2020)
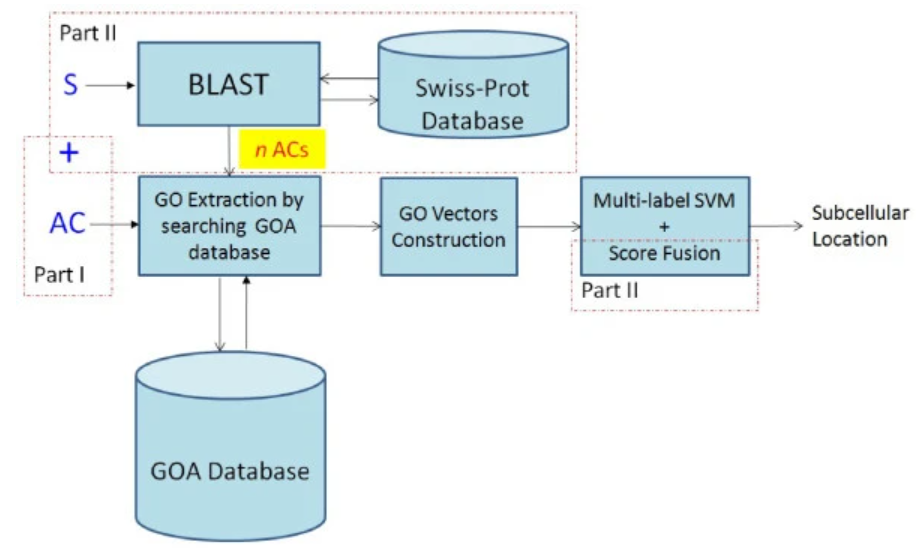
We present an interpretable and efficient web-server, namely FUEL-mLoc, using Feature-Unified prediction and Explanation of multi-Localization of cellular proteins in multiple organisms. This paper proposes a multi-label predictor based on ensemble linear neighborhood propagation (LNP), namely, LNP-Chlo, which leverages hybrid sequence-based feature information from both labeled and unlabeled proteins for predicting localization of both single- and multi-label chloroplast proteins.
S. Wan*, M.W. Mak* and S.Y. Kung
Bioinformatics 33 (5), 749-750 (2017)
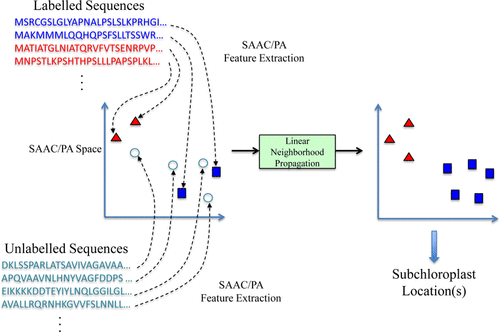
This paper proposes a multi-label predictor based on ensemble linear neighborhood propagation (LNP), LNP-Chlo, which leverages hybrid sequence-based feature information from both labeled and unlabeled proteins for predicting localization of both single- and multi-label chloroplast proteins. Experimental results on a stringent benchmark dataset and a novel independent dataset suggest that LNP-Chlo performs at least 6% (absolute) better than SOTA predictors. This paper also demonstrates that ensemble LNP significantly outperforms LNP based on individual features.
S. Wan*, M.W. Mak* and S.Y. Kung
Journal of Proteome Research 15 (12), 4755-4762 (2016)

This paper proposes an efficient predictor, namely Mem-mEN, which can produce sparse and interpretable solutions for predicting membrane proteins with single- and multi-label functional types.
S. Wan*, M.W. Mak* and S.Y. Kung*
IEEE/ACM Transactions on Computational Biology and Bioinformatics 13(4), 706–718 (2016)
We proposes an efficient multi-label predictor, mGOASVM, for predicting the subcellular localization of multi-location proteins. mGOASVM achieves an actual accuracy of 88.9% and 87.4%, respectively, which are significantly higher than those achieved by the SOTA predictors such as iLoc-Virus and iLoc-Plant.
S. Wan, M.W. Mak* and S.Y. Kung
BMC Bioinformatics 13, 1-16 (2012)
Full List of publications
Books
-
Machine learning for protein subcellular localization prediction
S. Wan and MW. Mak
De Gruyter, ISBN 978-1-5015-0150-0, 2015, Germany -
Bioinformatics and machine learning for cancer biology
S. Wan, Y. Fan, C. Jiang and S. Li
MDPI, ISBN 978-3-0365-4814-2, 2022, Switzerland. (edited book)
Journal Articles (*: corresponding author; #: co-first author; underlined names indicate lab members)
-
RaMBat: Accurate identification of medulloblastoma subtypes from diverse data sources with severe batch effects
M. Sun, J. Wang and S. Wan*
Molecular Oncology (2026) -
MetaPaCS: A novel meta-learning framework for pancreatic cancer subtype identification
N. Peterson, M. Sun, X. Wu, J. Wang and S. Wan*
bioRxiv 2025.12.29.696875 (2025) -
MOTLAB: A Weighted Multi-Omics Transfer Learning Approach to Mitigate Breast Cancer Racial Disparities
M. Baek, L. Li, V. Band, J. Wang and S. Wan*
bioRxiv 2025.12.22.696111 (2025) -
A Multi-Modal Transfer Learning Framework to Reduce Health Disparities in Prostate Adenocarcinoma
L. Li, J. Wang and S. Wan*
bioRxiv 2025.12.15.694538 (2025) -
Mapping Structural and Molecular Spatiotemporal Dynamics of Macaque Brain during Development and Aging
Y. Ma#, X. Wu#, R. Zhu#, Q. Sun#, F. Yang#, Z. Wang#, S. Dai#, X. Qiu, X. Yu, W. Sun, K. Chen, H. Wang, L. Cheng, S. Xie, T. Xu, Y. Chen, W. Ji*, S. Wan* and J. Wang*
Research Square -
RanBALL: An Ensemble Random Projection Model for Accurate Subtype Identification of Pediatric B-Cell Acute Lymphoblastic Leukemia
L. Li, H. Xiao, X. Wu, Z. Tang, J. Khoury, J. Wang and S. Wan*
Advanced Intelligent Systems, e202500965 (2025) -
Editorial: Subjective well-being and human decision behaviors
TJ. Huggins, DJ. Chai, S. Wan and CF. Lei
Frontiers in Psychology, Volume 16 - 2025 -
RPSLearner: A Novel Approach Based on Random Projection and Deep Stacking Learning for Categorizing Non-Small Cell Lung Cancer
X. Wu, J. Wang and S. Wan*
Advanced Intelligent Systems, e202500635 (2025) -
ECD co-operates with ERBB2 to promote tumorigenesis through upregulation of unfolded protein response and glycolysis
B.B. Kennedy#, M. Raza#, S. Mirza, A.R. Rajan, F. Oruji, M.M. Storck, S.M. Lele, T.E. Reznicek, L. Li, M.J. Rowley, S. Wan, B.C. Mohapatra*, H. Band*, V. Band*
Cancer Letters, 217959 (2025) -
AttentionAML: An attention-based deep learning framework for accurate molecular categorization of acute myeloid leukemia
L. Li,J. Khoury, J. Wang and S. Wan*
bioRxiv 2025.12.15.694538 (2025) -
The context-dependent epigenetic and organogenesis programs determine 3D vs. 2D cellular fitness of MYC-driven murine liver cancer cells
J. Fang, S. Singh, B. Wells, Q. Wu, H. Jin, L. Janke, S. Wan, J. Steele, J. Connelly, A. Murphy, R. Wang, A. Davidoff, M. Ashcroft, S. Pruett-Miller and J. Yang*
eLife 14:RP101299 (2025) -
A comprehensive review on RNA subcellular localization prediction
C. Zhang, X. Zhu, N. Peterson, J. Wang and S. Wan*
arXiv arXiv:2504.17162 (2025) -
Twelve Practical Recommendations for Developing and Applying Clinical Predictive Models
G. Feng*., H. Xu, S. Wan, H. Wang, X. Chen, R. Magari, Y. Han, Y. Wei and H. Gu
The Innovation Medicine 2(4), 100105 (2024) -
SAMP: Identifying antimicrobial peptides by an ensemble learning model based on proportionalized split amino acid composition
J. Feng#, M. Sun#, C. Liu, W. Zhang, C. Xu, J. Wang, G. Wang and S. Wan*
Briefings in Functional Genomics, 23(6), 879–890 (2024) -
Functional Connectivity Alterations in Cocaine Use Disorder: Insights from the Triple Network Model and the Addictions Neuroclinical Assessment Framework
Z. Xu, R. Liu, M. Azzam, S. Wan and J. Wang*
bioRxiv 2024.11.12.623073 (2024) -
A review of artificial intelligence-based brain age estimation and its applications for related diseases
M. Azzam, Z. Xu, R. Liu, L. Li, K.M. Soh, K.B. Challagundla, S. Wan and J. Wang*
Briefings in Functional Genomics, elae042 (2024) -
WIMOAD: Weighted integration of multi-omics data for Alzheimer’s Disease (AD) diagnosis
H. Xiao, J. Wang and S. Wan*
bioRxiv 2024.09.25.614862 (2024) -
RanBALL: An Ensemble Random Projection Model for Identifying Subtypes of B-cell Acute Lymphoblastic Leukemia
L. Li, H. Xiao, X. Wu, Z. Tang, J. Khoury, J. Wang and S. Wan*
bioRxiv 2024.09.24.614777 (2024) -
A prognostic framework for predicting lung signet ring cell carcinoma via a machine learning based cox proportional hazard model
H. Chen, Y. Xu, H. Lin, S. Wan* and L. Luo*
Journal of Cancer Research and Clinical Oncology 150(364), 1-15 (2024) -
Multi-Omics based artificial intelligence for cancer research
L. Li#, M. Sun#, J. Wang and S. Wan*
Advances in Cancer Research 163, 303-356 (2024) -
Artificial intelligence for omics data analysis
Z. Ahmed#, S. Wan#, F. Zhang# and W. Zhong#
BMC Methods 1, 4 (2024) -
A review for artificial intelligence based protein subcellular localization
H. Xiao, Y. Zou, J. Wang and S. Wan*
Biomolecules 14, 409 (2024) -
Procyanidin alleviates ferroptosis and inflammation of LPS-induced RAW264.7 cell via the Nrf2/HO-1 pathway
J. Zeng, Y. Weng, T. Lai, L. Chen, Y. Li, Q. Huang, S. Zhong, S. Wan* and L. Luo*
Naunyn-Schmiedeberg’s Arch Pharmacol (2023) -
Editorial: Bioinformatics analysis of omics data for biomarker identification in clinical research, Volume II
M. Sun#, L. Li#, H. Xiao#, J. Feng#, J. Wang and S. Wan*
Frontiers in Genetics 14, 1256468 (2023) -
USP1 expression driven by EWS:: FLI1 transcription factor stabilizes Survivin and mitigates replication stress in Ewing sarcoma
H. J. Mallard, S. Wan, P. Nidhi, Y. D. Hanscom-Trofy, B. Mohapatra, N. T. Woods, J. A. Lopez-Guerrero, A. Llombart-Bosch, I. Machado, K. Scotlandi, N. F. Kreiling, M. C. Perry, S. Mirza, D. W. Coulter, V. Band, H. Band and G. Ghosal
Molecular Cancer Research MCR, MCR-23-0323 (2023) -
Embedded bioprinting of breast tumor cells and organoids using low concentration collagen based bioinks
W. Shi, S. Mirza, M. Kuss, B. Liu, A. Hartin, S. Wan, Y. Kong, B. Mohapatra, M. Krishnan, H. Band, V. Band and B. Duan
Advanced Healthcare Materials e2300905 (2023) -
Editorial: Ferroptosis as a novel therapeutic target for inflammation-related diseases
Y. Liang, Z. Su, X. Mao, S. Wan* and L. Luo*
Frontiers in Pharmacology 14, 1152326 (2023) -
Editorial: Single cell meets metabolism and cancer biology
J. Wang and S. Wan*
Frontiers in Oncology 13, 1125186 (2023) -
Etiology of oncogenic fusions in 5,190 childhood cancers and its clinical and therapeutic implication
Y. Liu, J. Klein, R. Bajpai, L. Dong, Q. Tran, P. Kolekar, J. L. Smith, R. E. Ries, B. J. Huang, Y. C. Wang, T. A. Alonzo, L. Tian, H. L. Mulder, T. I. Shaw, J. Ma, M. P. Walsh, G. Song, T. Westover, R. J. Autry, A. M. Gout, D. A. Wheeler, S. Wan, G. Wu, J. J. Yang, W. E. Evans, M. Loh, J. Easton, J. H. Zhang, J. M. Klco and X. Ma
Nature Communications 14 (1), 1739 (2023) -
The nuclear receptor ERR cooperates with the cardiogenic factor GATA4 to orchestrate transcriptional control of cardiomyocyte differentiation
T. Sakamoto, K. Batmanov, S. Wan, Y. Guo, L. Lai, R. B. Vega and D. P. Kelly
Nature Communications 13, 1991 (2022) -
Alzheimer’s disease-associated U1 snRNP splicing dysfunction causes neuronal hyperexcitability and cognitive impairment
P. C. Chen, X. Han, T. I. Shaw, Y. Fu, H. Sun, M. Niu, Z. Wang, Y. Jiao, B. J. W. Teubner, D. Eddins, L. N. Beloate, B. Bai, J. Mertz, Y. Li, J. H. Cho, X. Wang, Z. Wu, D. Liu, S. Poudel, Z. F. Yuan, A. Mancieri, J. Low, H. M. Lee, M. Patton, L. Earls, E. Stewart, P. Vogel, S. Wan, G. Serrano, T. Beach, M. Dyer, R. Smeyne, T. Moldoveanu, T. Chen, G. Wu, S. Zakharenko, G. Yu and J. Peng
Nature Aging 2(10), 923–940 (2022) -
A sequence obfuscation method for protecting personal genomic privacy
S. Wan* and J. Wang*
Frontiers in Genetics 13, 876686 (2022) -
Genomic profiling identifies genes and pathways dysregulated by HEY1–NCOA2 fusion and shines a light on mesenchymal chondrosarcoma tumorigenesis
W. Qi, W. Rosikiewicz, Z. Yin, B. Xu, H. Jiang, S. Wan, Y. Fan, G. Wu and L. Wang
The Journal of Pathology 257 (5), 579-592 (2022) -
Special issue on bioinformatics and machine learning for cancer biology
S. Wan*, C. Jiang, S. Li and Y. Fan
Biology 11 (3), 361 (2022) -
Identification of a modular super-enhancer in murine retinal development
V. Honnell, J. L. Norrie, A. G. Patel, C. Ramirez, J. Zhang, Y. H. Lai, S. Wan and M. A. Dyer
Nature Communications 13 (1), 253 (2022) -
Improving bulk RNA-seq classification by transferring gene signature from single cells in acute myeloid leukemia
R. Wang, X. Zheng, J. Wang, S. Wan, F. Song, M. H. Wong, K. S. Leung and L. Cheng
Briefings in Bioinformatics 23 (2), bbac002 (2022) -
Editorial: Transcriptional regulation in metabolism and immunology
C. Jiang, S. Wan, P. Hu, Y. Li and S. Li
Frontiers in Genetics 13, 845697 (2022) -
Targeting the spliceosome through RBM39 degradation results in exceptional responses in high-risk neuroblastoma models
S. Singh, W. Quarni, M. Goralski, S. Wan, H. Jin, L. A. Van de Velde, J. Fang, Q. Wu, A. Abu-Zaid, T. Wang, R. Singh, D. Craft, Y. Fan, T. Confer, M. Johnson, W. J. Akers, R. Wang, P. J. Murray, P. G. Thomas, D. Nijhawan, A. M. Davidoff and J. Yang
Science Advances 7 (47), eabj5405 (2021) -
YAP/TAZ maintain the proliferative capacity and structural organization of radial glial cells during brain development
A. Lavado, R. Gangwar, J. Paré, S. Wan, Y. Fan and X. Cao
Developmental Biology 480, 39-49 (2021) -
SHARP: hyperfast and accurate processing of single-cell RNA-seq data via ensemble random projection
S. Wan, J. Kim and K. J. Won
Genome Research 30 (2), 205-213 (2020) -
A critical role for estrogen-related receptor signaling in cardiac maturation
T. Sakamoto, T. R. Matsuura, S. Wan, D. M. Ryba, J. U. Kim, K. J. Won, L. Lai, C. Petucci, N. Petrenko, K. Musunuru, R. B. Vega and D. P. Kelly
Circulation Research 126 (12), 1685-1702 (2020) -
MondoA drives muscle lipid accumulation and insulin resistance
B. Ahn, S. Wan, N. Jaiswal, R. B. Vega, D. E. Ayer, P. M. Titchenell, X. Han, K. J. Won and D. P. Kelly
JCI Insight 4 (15) (2019) -
Predicting subcellular localization of multi-location proteins by improving support vector machines with adaptive-decision schemes
S. Wan* and M.W. Mak*
International Journal of Machine Learning and Cybernetics 9, 399-411 (2018) -
Is congenital amusia a disconnection syndrome? A study combining tract-and network-based analysis
J. Wang, C. Zhang, S. Wan and G. Peng
Frontiers in Human Neuroscience 11, 473 (2017) -
Gram-LocEN: Interpretable prediction of subcellular multi-localization of Gram-positive and Gram-negative bacterial proteins
S. Wan*, M.W. Mak* and S.Y. Kung
Chemometrics and Intelligent Laboratory Systems 162, 1-9 (2017) -
FUEL-mLoc: feature-unified prediction and explanation of multi-localization of cellular proteins in multiple organisms
S. Wan*, M.W. Mak* and S.Y. Kung
Bioinformatics 33 (5), 749-750 (2017) -
Transductive learning for multi-label protein subchloroplast localization prediction
S. Wan*, M.W. Mak* and S.Y. Kung*
IEEE/ACM Transactions on Computational Biology and Bioinformatics 14(1), 212–224 (2017) -
Ensemble linear neighborhood propagation for predicting subchloroplast localization of multi-location proteins
S. Wan*, M.W. Mak* and S.Y. Kung
Journal of Proteome Research 15 (12), 4755-4762 (2016) -
Benchmark data for identifying multi-functional types of membrane proteins
S. Wan*, M.W. Mak* and S.Y. Kung
Data in Brief 8, 105-107 (2016) -
Mem-ADSVM: A two-layer multi-label predictor for identifying multi-functional types of membrane proteins
S. Wan*, M.W. Mak* and S.Y. Kung
Journal of Theoretical Biology 398, 32-42 (2016) -
Sparse regressions for predicting and interpreting subcellular localization of multi-label proteins
S. Wan*, M.W. Mak* and S.Y. Kung
BMC Bioinformatics 17 (1), 1-17 (2016) -
Mem-mEN: predicting multi-functional types of membrane proteins by interpretable elastic nets
S. Wan*, M.W. Mak* and S.Y. Kung*
IEEE/ACM Transactions on Computational Biology and Bioinformatics 13(4), 706–718 (2016) -
mLASSO-Hum: a LASSO-based interpretable human-protein subcellular localization predictor
S. Wan*, M.W. Mak and S.Y. Kung
Journal of Theoretical Biology 382, 223-234 (2015) -
mPLR-Loc: An adaptive-decision multi-label classifier based on penalized logistic regression for protein subcellular localization prediction
S. Wan, M.W. Mak and S.Y. Kung
Analytical Biochemistry 473, 14-27 (2015) -
R3P-Loc: A compact multi-label predictor using ridge regression and random projection for protein subcellular localization
S. Wan, M.W. Mak and S.Y. Kung
Journal of Theoretical Biology 360, 34-45 (2014) -
HybridGO-Loc: mining hybrid features on gene ontology for predicting subcellular localization of multi-location proteins
S. Wan, M.W. Mak and S.Y. Kung
PloS One 9 (3), e89545 (2014) -
Semantic similarity over gene ontology for multi-label protein subcellular localization
S. Wan*, M.W. Mak* and S.Y. Kung*
Engineering 5 (10), 68 (2013) -
GOASVM: A subcellular location predictor by incorporating term-frequency gene ontology into the general form of Chou’s pseudo-amino acid composition
S. Wan*, M.W. Mak* and S.Y. Kung*
Journal of Theoretical Biology 323, 40-48 (2013) -
mGOASVM: Multi-label protein subcellular localization based on gene ontology and support vector machines
S. Wan, M.W. Mak and S.Y. Kung
BMC Bioinformatics 13, 1-16 (2012)
Conference Abstracts
-
Exploring high-resolution subtypes for pancreatic ductal adenocarcinoma via a meta-clustering approach
N. Peterson, J. Wang and S. Wan*.
Cancer Research, 2026 -
DAG-guided kernel learning framework to mitigate racial disparities in breast cancer
M. Baek, J. Wang, V. Band and S. Wan*.
Cancer Research, 2026 -
OTTER: Optimal transport-based transcriptomics and genomics representation fusion for T-ALL subtyping
L. Li, J. Wang and S. Wan*.
Cancer Research, 2026 -
Integrated Meta-Clustering Reveals Subtype-Specific Transcriptomic Heterogeneity of Pediatric Low-Grade Glioma
B. Tuerhanbayi, J. Wang and S. Wan*.
Cancer Research, 2026 -
iS3RGs: Discriminating diverse medulloblastoma subtypes by leveraging heterogenous transcriptome data with batch effects
M. Sun, J. Wang and S. Wan*.
Cancer Research, 2025, vol. 85 (8_Supplement_1), pp. 5022-5022. (presented in AACR Annual Meeting 2025, Chicago, IL, Apr. 2025) -
AttentionAML: An attention-based deep learning model for accurate identification of childhood acute myeloid leukemia subtype
L. Li, J. Khoury, J. Wang and S. Wan*.
Cancer Research, 2025, vol. 85 (8_Supplement_1), pp. 7425-7425. (presented in AACR Annual Meeting 2025, Chicago, IL, Apr. 2025) -
Identifying pancreatic cancer subtypes by a novel meta-learning model
N. Peterson, M. Sun, J. Wang and S. Wan*.
Cancer Research, 2025, vol. 85 (8_Supplement_1), pp. 1104-1104. (presented in AACR Annual Meeting 2025, Chicago, IL, Apr. 2025) -
Combining random projection and stacking learning for NSCLC subtype classification based on transcriptomic data
X. Wu, J. Wang and S. Wan*.
Cancer Research, 2025, vol. 85 (8_Supplement_1), pp. 1098-1098. (presented in AACR Annual Meeting 2025, Chicago, IL, Apr. 2025) -
A weighted multi-modal transfer learning model for alleviating racial disparities in breast cancer
M. Baek, L. Li, J. Wang, V. Band and S. Wan*.
Cancer Research, 2025, vol. 85 (8_Supplement_1), pp. 3620-3620. (presented in AACR Annual Meeting 2025, Chicago, IL, Apr. 2025) -
ECD co-operates with ERBB2 to promote tumorigenesis through upregulation of unfolded protein response and glycolysis
B. Kennedy, M. Raza, S. Mirza, A. Rajan, F. Oruji, Ma. Storck, S. Lele, T. Reznicek, L. Li, J. Rowley, S. Wan, B. Mohapatra, H. Band and V. Band*.
AACR Annual Meeting 2025, Chicago, IL, Apr. 2025 -
Targeting CBL and CBLB ubiquitin ligases to exhaust cancer stem cells in metastatic breast cancer
B. Mohapatra, A. Bhat, M. Raza, H. Luan, S. Shrestha, S. Kolluru, M. Storck, L. Li, F. Kuo, S. Leit, F. Qiu, S. Lele, D. Ciccone, C. Loh, S. Wan, V. Band and H. Band*.
AACR Annual Meeting 2025, Chicago, IL, Apr. 2025 -
Enhancing prostate pelvic multimodality data generating with conditional generative models: A Pix2Pix-based approach for MRI-to-PET synthesis
R. Liu, S. Wang, S. Wan and J. Wang*.
AACR Annual Meeting 2025, Chicago, IL, Apr. 2025 -
Development of g-rate based random forest machine learning model to predict overall survival for patients with metastatic prostate cancer
M. Azzam, H. Leuva, M. Zhou, B. Teply, R. Bergan, S. Bates, S. Wan, A. Fojo and J. Wang*.
AACR Annual Meeting 2025, Chicago, IL, Apr. 2025 -
Identifying antimicrobial peptides based on proportionalized split pseudo amino acid compositions
M. Sun, J. Wang and S. Wan*.
International Conference on Intelligent Biology and Medicine (ICIBM 2024), Houston, TX, Sep. 2024 -
Discriminating diverse medulloblastoma subtypes by leveraging heterogenous transcriptome data with batch effects
M. Sun, J. Wang and S. Wan*.
International Conference on Intelligent Biology and Medicine (ICIBM 2024), Houston, TX, Sep. 2024 -
Pediatric acute myeloid leukemia classification via multi-modal ensemble learning
L. Li, J. Khoury, J. Wang and S. Wan*.
International Conference on Intelligent Biology and Medicine (ICIBM 2024), Houston, TX, Sep. 2024 -
WIMOAD: Weighted integration of multi-omics data for Alzheimer’s Disease (AD) diagnosis
H. Xiao, J. Wang and S. Wan*.
International Conference on Intelligent Biology and Medicine (ICIBM 2024), Houston, TX, Sep. 2024 -
iS3RGs: Discriminating diverse medulloblastoma subtypes by leveraging heterogenous transcriptome data with batch effects
M. Sun, J. Wang and S. Wan*.
Pediatric Cancer Research Symposium, Omaha, NE, Aug. 2024 -
AttentionAML: An attention-based deep learning model for identifying acute myeloid leukemia subtypes
L. Li, J. Khoury, J. Wang and S. Wan*.
Pediatric Cancer Research Symposium, Omaha, NE, Aug. 2024 -
RanBALL: Identifying B-cell acute lymphoblastic leukemia subtypes based on an ensemble random projection model, Cancer Research, vol. 84 (6_ Supplement), pp.4907-4907.
L. Li, H. Xiao, J. Khoury, J. Wang and S. Wan*.
AACR Annual Meeting 2024, San Diego, CA, Apr. 2024 -
Reducing health disparities for prostate adenocarcinoma by integrating multi-omics data via a multi-modal transfer learning approach, Cancer Research, vol. 84 (6_ Supplement), pp.4800-4800.
L. Li, J. Wang and S. Wan*.
AACR Annual Meeting 2024, San Diego, CA, Apr. 2024 -
SAMP: An accurate ensemble model based on proportionalized split amino acid composition for identifying antimicrobial peptides
J. Feng, M. Sun, W. Zhang, G. Wang and S. Wan*.
Antimicrobial Peptides, Yesterday, Today and Tomorrow 2023, Omaha, NE, Oct (2023) -
B-cell acute lymphoblastic leukemia subtype identification with an ensemble random projection-based machine learning model
L. Li, H. Xiao and S. Wan.
CHRI Scientific Conference 2023, Omaha, NE, Nov (2023) -
Integrating multi-omics data by a multi-modal transfer learning model to reduce healthcare disparities for kidney renal clear cell carcinoma
L. Li and S. Wan.
CHRI Scientific Conference 2023, Omaha, NE, Nov (2023) -
RanBall: An ensemble random projection-based model for identifying B-cell acute lymphoblastic leukemia subtypes
L. Li, H. Xiao and S. Wan.
PCRG symposium 2023, Omaha, NE, Aug (2023) -
RNA-seq and chIP-seq profiling identifies genes and pathways dysregulated by hey1-ncoa2 fusion and shed a light on mesenchymal chondrosarcoma tumorigenesis
W. Qi, W. Rosikiewicz, Z. Yin, B. Xu, S. Wan, Y. Fan, G. Wu and L. Wang.
AACR Annual Meeting 2021, Philadelphia, PA, Apr (2021) -
The estrogen-related receptor (ERR) drives cardiac myocyte maturation in cooperation with GATA4
T. Sakamoto, S. Wan, K. Batmanov and D.P. Kelly.
Circulation Research, 127(A222-A222) (2020) -
Hyper-fast and accurate clustering of ultra-large-scale single-cell data with ensemble random projection
S. Wan, J. Kim, Y. Fan and K.J. Won.
Cell Symposia, The Conceptual Power of Single-Cell Biology, San Francisco, CA, USA, Apr (2020). (postponed due to COVID-19 outbreak) -
The estrogen-related receptor coordinates transcription of genes involved in mitochondrial and contractile maturation in human induced pluripotent stem cell-derived cardiac myocytes
T. Sakamoto, S. Wan, K.J. Won and D.P. Kelly.
Circulation, vol. 140 (Suppl_1), pp. A11803-A11803 (2019). (presented in American Heart Association Scientific Session (AHA2019), Philadelphia, PA, USA, Nov (2019) -
Estrogen-related receptor signaling is critical for postnatal cardiac maturation
T.R. Matsuura, T. Sakamoto, D.M. Ryba, S. Wan and D.P. Kelly.
Circulation, vol. 140 (Suppl_1), pp. A11803-A11803 (2019). (presented in American Heart Association Scientific Session (AHA2019), Philadelphia, PA, USA) -
MondoA mediates myocyte lipid accumulation and insulin resistance driven by chronic nutrient excess
B. Ahn, S. Wan, K.J. Won, N. Jaiswal, P.M. Titchenell and D.P. Kelly.
American Diabetes Association’s 79th Scientific Sessions (ADA2019), San Francisco, CA, USA, Jun (2019) (oral) -
Hyper-fast and accurate processing of large-scale single-cell transcriptomics data via ensemble random projection
S. Wan, J. Kim and K.J. Won.
RECOMB/ISCB Conference on Regulatory & Systems Genomics with DREAM Challenges (RSG DREAM 2018), New York, USA, Dec (2018)
Conference Papers
-
nnU-BNST: Deep Learning-Based Automated Segmentation of the Bed Nucleus of the Stria Terminalis
Z. Xu, M. Azzam, M. Bivens, M.Dustin, J.U. Blackford, S. Wan and J. Wang*
Machine Learning in Medical Imaging. MLMI 2025. Lecture Notes in Computer Science, vol 16241. -
Processing millions of single cells by SHARP
S. Wan, J. Kim and K.J. Won.
The 11th ACM Conference on Bioinformatics, Computational Biology and Health Informatics (ACM BCB 2020), virtual online, Sep (2020) -
Hyper-fast and accurate clustering of ultra-large-scale single-cell data with ensemble random projection
S. Wan, J. Kim, Y. Fan and K.J. Won.
The 2020 International Conference on Machine Learning (ICML) Workshop on Computational Biology, virtual online, Jul (2020) -
Protecting genomic privacy by a sequence-similarity based obfuscation method
S. Wan, M.W. Mak and S.Y. Kung.
2017, arXiv preprint arXiv, 1708.02629 (2017) -
Ratio utility and cost analysis for privacy preserving subspace projection
S. Wan and S.Y. Kung.
2017, arXiv preprint arXiv, 1702.07976 (2017) -
Ensemble random projection for multi-label classification with application to protein subcellular localization
S. Wan, M.W. Mak, B. Zhang, Y. Wang and S.Y. Kung.
2014 IEEE International Conference on Acoustic Speech and Signal Processing (ICASSP’14), Florence, Italy, May 2014, pp. 5999-6003 (2014) -
An ensemble classifier with random projection for predicting multi-label protein subcellular localization
S. Wan, M.W. Mak, B. Zhang, Y. Wang and S.Y. Kung.
The 2013 IEEE International Conference on Bioinformatics and Biomedicine (BIBM’2013), Shanghai, China, Dec. 2013, pp. 35-42 (2013) -
Adaptive thresholding for multi-label SVM classification with application to protein subcellular localization prediction
S. Wan, M.W. Mak and S.Y. Kung.
2013 IEEE International Conference on Acoustic Speech and Signal Processing (ICASSP’13), Vancouver, Canada, May 2013, pp. 3547-3551 (2013) -
GOASVM: Protein subcellular localization prediction based on Gene ontology annotation and SVM
S. Wan, M.W. Mak and S.Y. Kung.
2012 IEEE International Conference on Acoustic Speech and Signal Processing (ICASSP’12), Kyoto, Japan, Mar. 2012, pp. 2229-2232 (2012) -
Protein subcellular localization prediction based on profile alignment and Gene Ontology
S. Wan, M.W. Mak and S.Y. Kung.
2011 IEEE International Workshop on Machine Learning for Signal Processing (MLSP’11), Beijing, China, Sep. 2011, pp. 1-6 (2011) -
A method of continuous data flow embedded within speech signals
S. Wan, C. Yao, Y. Hu and G. Zhang.
The 2-nd International Conference on Signal Acquisition and Processing (ICSAP’10), Bangalore, India, Feb. 2010, pp. 362-365 (2010)